NYT COVID Forecast Aggregation (Box-Cox)
New York Times Data on COVID
I will grab the dataset from the NYT COVID data github. I will create two variables, New Cases and New Deaths to model. The final line use aggregation to create the national data.
NYT.COVIDN <- read.csv(url("https://raw.githubusercontent.com/nytimes/covid-19-data/master/us-states.csv"))
# Define a tsibble; the date is imported as character so mutate that first.
NYT.COVID <- NYT.COVIDN %>%
mutate(date = as.Date(date)) %>%
as_tsibble(index = date, key = state) %>%
group_by(state) %>%
mutate(New.Cases = difference(cases), New.Deaths = difference(deaths)) %>%
filter(!(state %in% c("Guam", "Puerto Rico", "Virgin Islands", "Northern Mariana Islands")))
NYT.COVID <- NYT.COVID %>%
mutate(New.Cases = (New.Cases >= 0) * New.Cases, New.Deaths = (New.Deaths >=
0) * New.Deaths)
NYTAgg.COVID <- NYT.COVID %>%
aggregate_key(state, New.Cases = sum(New.Cases, na.rm = TRUE), New.Deaths = sum(New.Deaths,
na.rm = TRUE)) %>%
filter(date > as.Date("2020-03-31")) %>%
mutate(Day.of.Week = as.factor(wday(date, label = TRUE)))
lambdaC <- NYTAgg.COVID %>%
filter(is_aggregated(state)) %>%
features(New.Cases, features = guerrero) %>%
pull(lambda_guerrero)
P1 <- NYTAgg.COVID %>%
filter(is_aggregated(state)) %>%
autoplot(New.Cases)
P2 <- NYTAgg.COVID %>%
filter(is_aggregated(state)) %>%
autoplot(box_cox(New.Cases, lambdaC)) + labs(y = "Box-Cox(New.Cases)")
library(patchwork)
P1/P2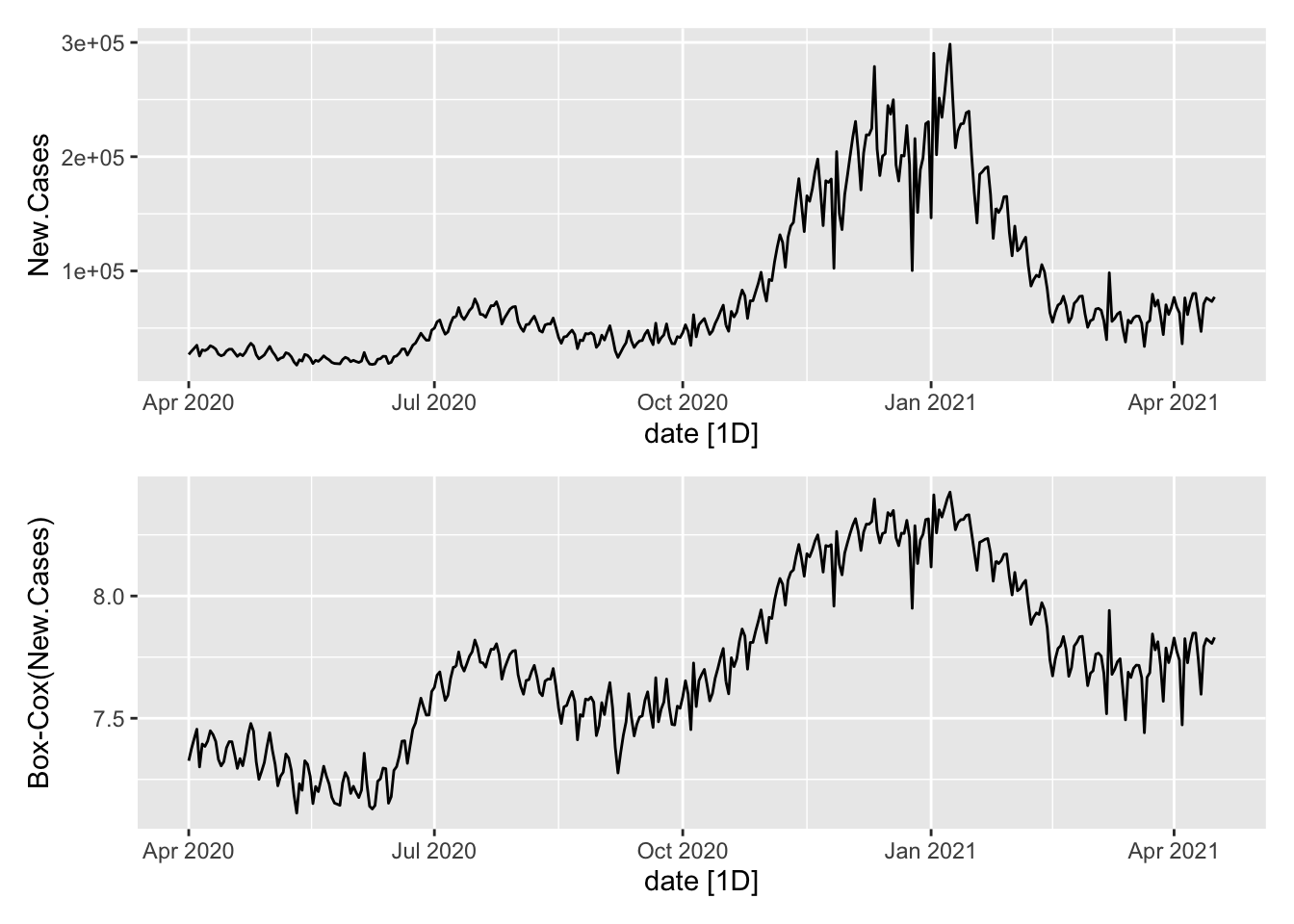
The \(\lambda\) for deaths…
lambdaD <- NYTAgg.COVID %>%
filter(is_aggregated(state)) %>%
features(New.Deaths, features = guerrero) %>%
pull(lambda_guerrero)
P1 <- NYTAgg.COVID %>%
filter(is_aggregated(state)) %>%
autoplot(New.Deaths)
P2 <- NYTAgg.COVID %>%
filter(is_aggregated(state)) %>%
autoplot(box_cox(New.Deaths, lambdaD)) + labs(y = "Box-Cox(New.Deaths)")
library(patchwork)
P1/P2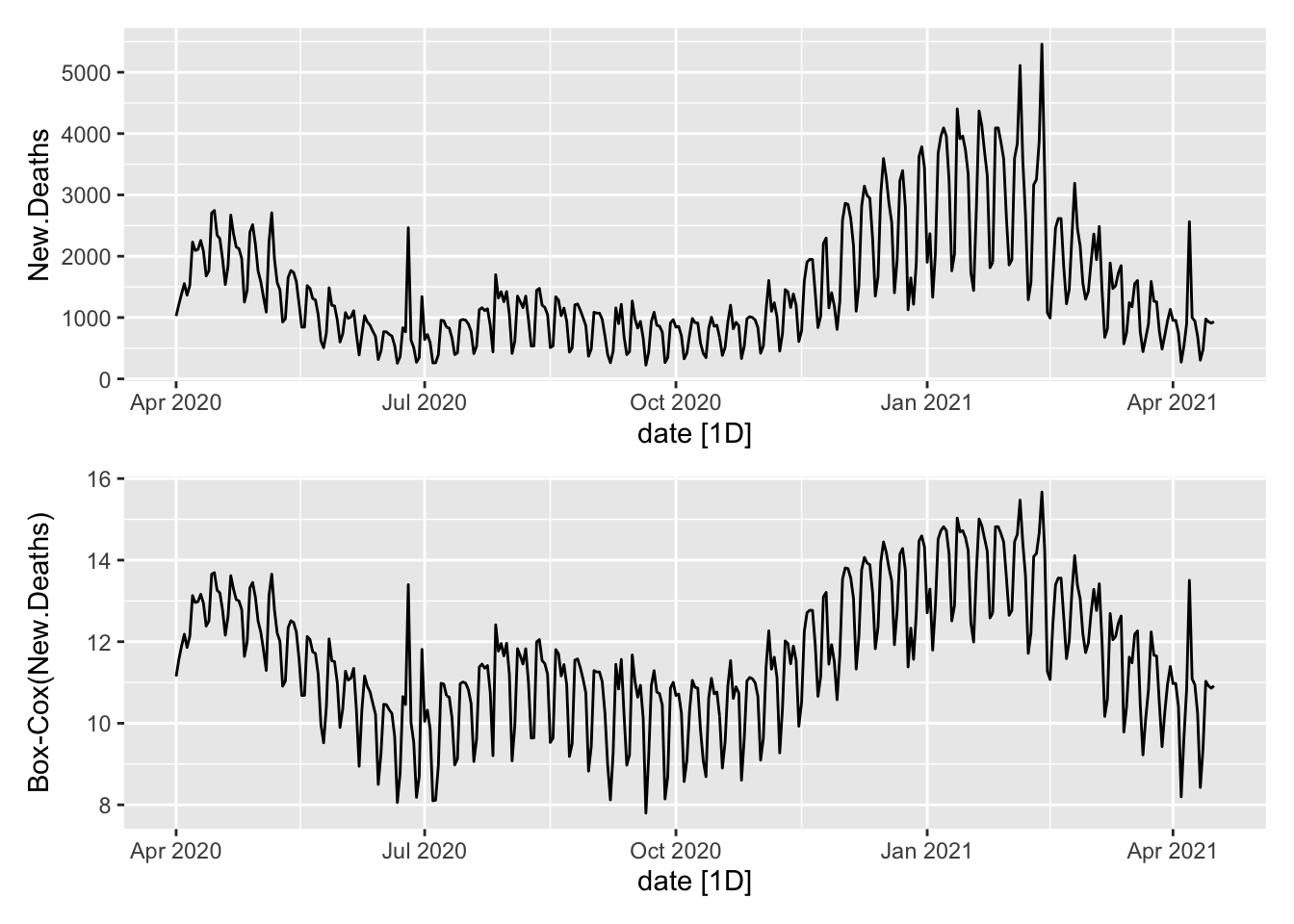
Plot the two aggregates.
plot1 <- NYTAgg.COVID %>%
filter(is_aggregated(state)) %>%
autoplot(box_cox(New.Cases, lambdaC)) + labs(y = "Box-Cox(New.Cases)")
plot2 <- NYTAgg.COVID %>%
filter(is_aggregated(state)) %>%
autoplot(box_cox(New.Deaths, lambdaD)) + labs(y = "Box-Cox(New.Deaths)")
plot1/plot2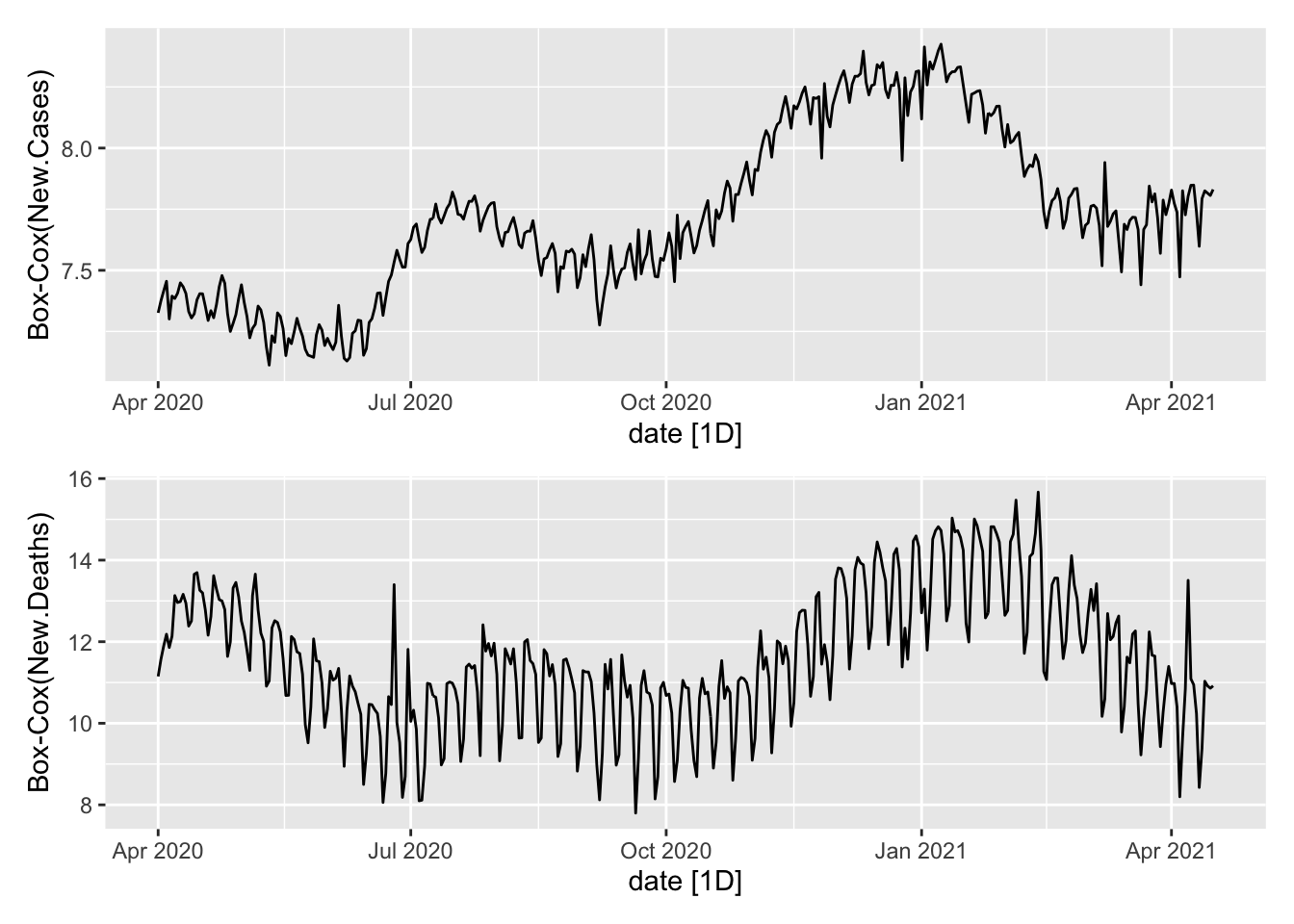
Are there day of week effects?
NYTAgg.COVID %>%
model(TSLM(box_cox(New.Cases, lambdaC) ~ Day.of.Week)) %>%
glance() %>%
filter(p_value < 0.05)# A tibble: 15 x 16
state .model r_squared adj_r_squared sigma2 statistic p_value df
<chr*> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <int>
1 Connecticu… TSLM(box… 0.0785 0.0592 0.742 4.06 6.31e- 4 7
2 District o… TSLM(box… 0.0409 0.0255 0.248 2.65 1.57e- 2 7
3 Georgia … TSLM(box… 0.0409 0.0255 0.235 2.65 1.57e- 2 7
4 Idaho … TSLM(box… 0.0610 0.0453 0.928 3.88 9.21e- 4 7
5 Iowa … TSLM(box… 0.0478 0.0325 0.306 3.13 5.26e- 3 7
6 Kansas … TSLM(box… 0.538 0.530 0.654 67.9 1.01e-55 7
7 Kentucky … TSLM(box… 0.0417 0.0263 0.669 2.71 1.37e- 2 7
8 Louisiana … TSLM(box… 0.0821 0.0649 0.282 4.78 1.09e- 4 7
9 Michigan … TSLM(box… 0.320 0.309 0.490 29.3 8.89e-29 7
10 Mississipp… TSLM(box… 0.0868 0.0719 0.287 5.84 7.75e- 6 7
11 Nebraska … TSLM(box… 0.0392 0.0238 0.368 2.54 2.03e- 2 7
12 Rhode Isla… TSLM(box… 0.0718 0.0513 0.463 3.51 2.33e- 3 7
13 South Dako… TSLM(box… 0.0672 0.0520 0.540 4.42 2.51e- 4 7
14 Washington… TSLM(box… 0.124 0.110 0.311 8.83 5.10e- 9 7
15 Wisconsin … TSLM(box… 0.0364 0.0208 0.427 2.34 3.13e- 2 7
# … with 8 more variables: log_lik <dbl>, AIC <dbl>, AICc <dbl>, BIC <dbl>,
# CV <dbl>, deviance <dbl>, df.residual <int>, rank <int>In 12 states, there appear to be and in a few of them, those effects are large. Modelling this covariate seems useful. At the same time, this does nothing about the broad trends.
NYTAgg.COVID %>%
model(TSLM(box_cox(New.Deaths, lambdaD) ~ Day.of.Week)) %>%
glance() %>%
filter(p_value < 0.05)# A tibble: 47 x 16
state .model r_squared adj_r_squared sigma2 statistic p_value df
<chr*> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <int>
1 Alabama TSLM(box… 0.278 0.267 12.4 24.0 4.80e-24 7
2 Alaska TSLM(box… 0.0711 0.0562 14.6 4.77 1.06e- 4 7
3 Arizona TSLM(box… 0.461 0.452 6.87 53.2 2.72e-47 7
4 Arkansas TSLM(box… 0.0454 0.0301 7.54 2.97 7.67e- 3 7
5 California TSLM(box… 0.187 0.174 2.89 14.3 1.09e-14 7
6 Colorado TSLM(box… 0.109 0.0948 9.33 7.63 9.59e- 8 7
7 Connecticut TSLM(box… 0.407 0.398 16.3 42.8 1.07e-39 7
8 Delaware TSLM(box… 0.0470 0.0317 16.5 3.07 6.00e- 3 7
9 Florida TSLM(box… 0.0749 0.0601 3.49 5.05 5.41e- 5 7
10 Georgia TSLM(box… 0.356 0.346 3.35 34.5 3.80e-33 7
# … with 37 more rows, and 8 more variables: log_lik <dbl>, AIC <dbl>,
# AICc <dbl>, BIC <dbl>, CV <dbl>, deviance <dbl>, df.residual <int>,
# rank <int>AN STL
NYTAgg.COVID %>%
filter(is_aggregated(state)) %>%
model(STLC = STL(box_cox(New.Cases, lambdaC) ~ trend(window = 14) + season(window = 21),
robust = TRUE)) %>%
components() %>%
autoplot()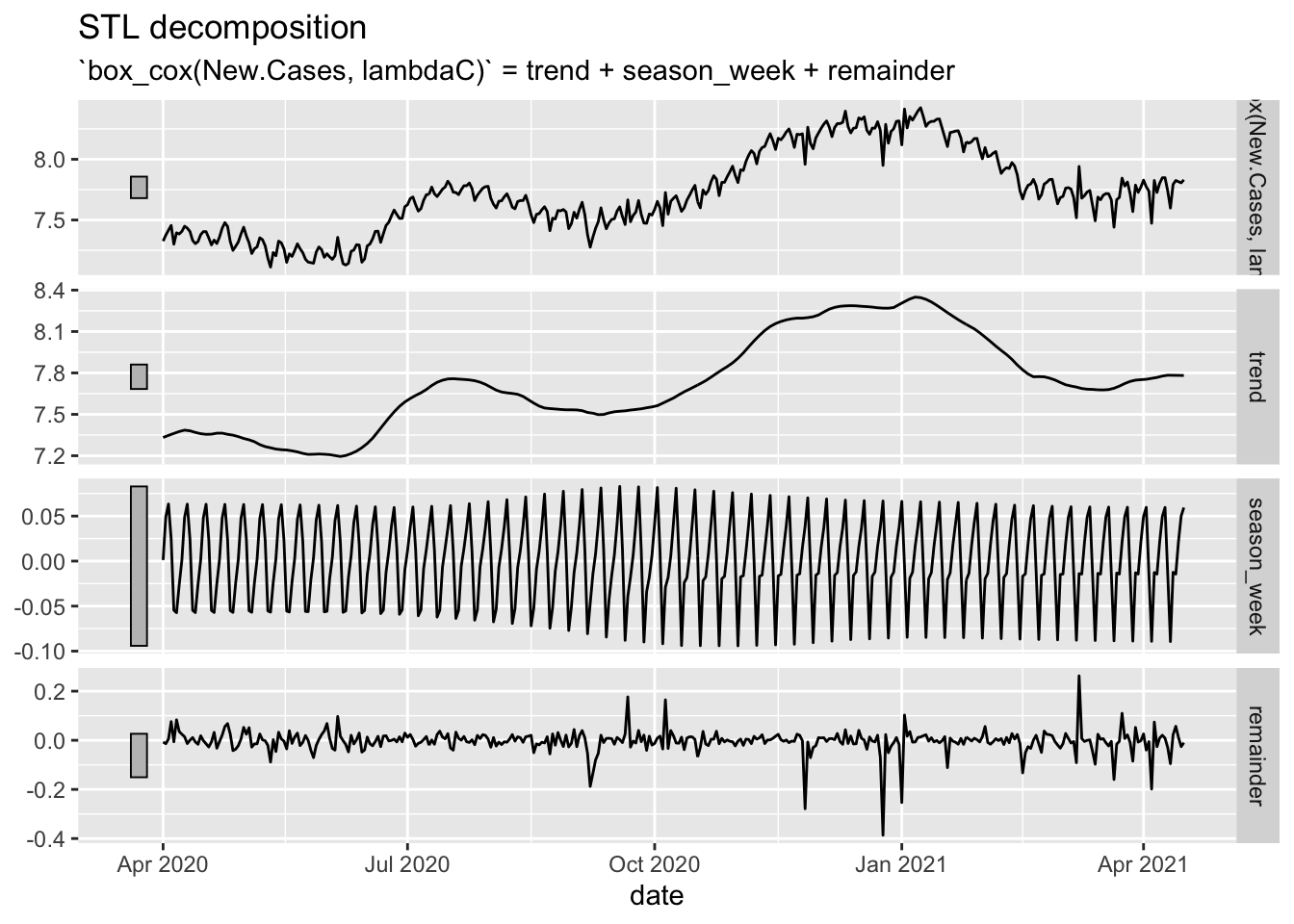
NYTAgg.COVID %>%
filter(is_aggregated(state)) %>%
model(STLC = STL(box_cox(New.Deaths, lambdaD) ~ trend(window = 14) + season(window = 21),
robust = TRUE)) %>%
components() %>%
autoplot()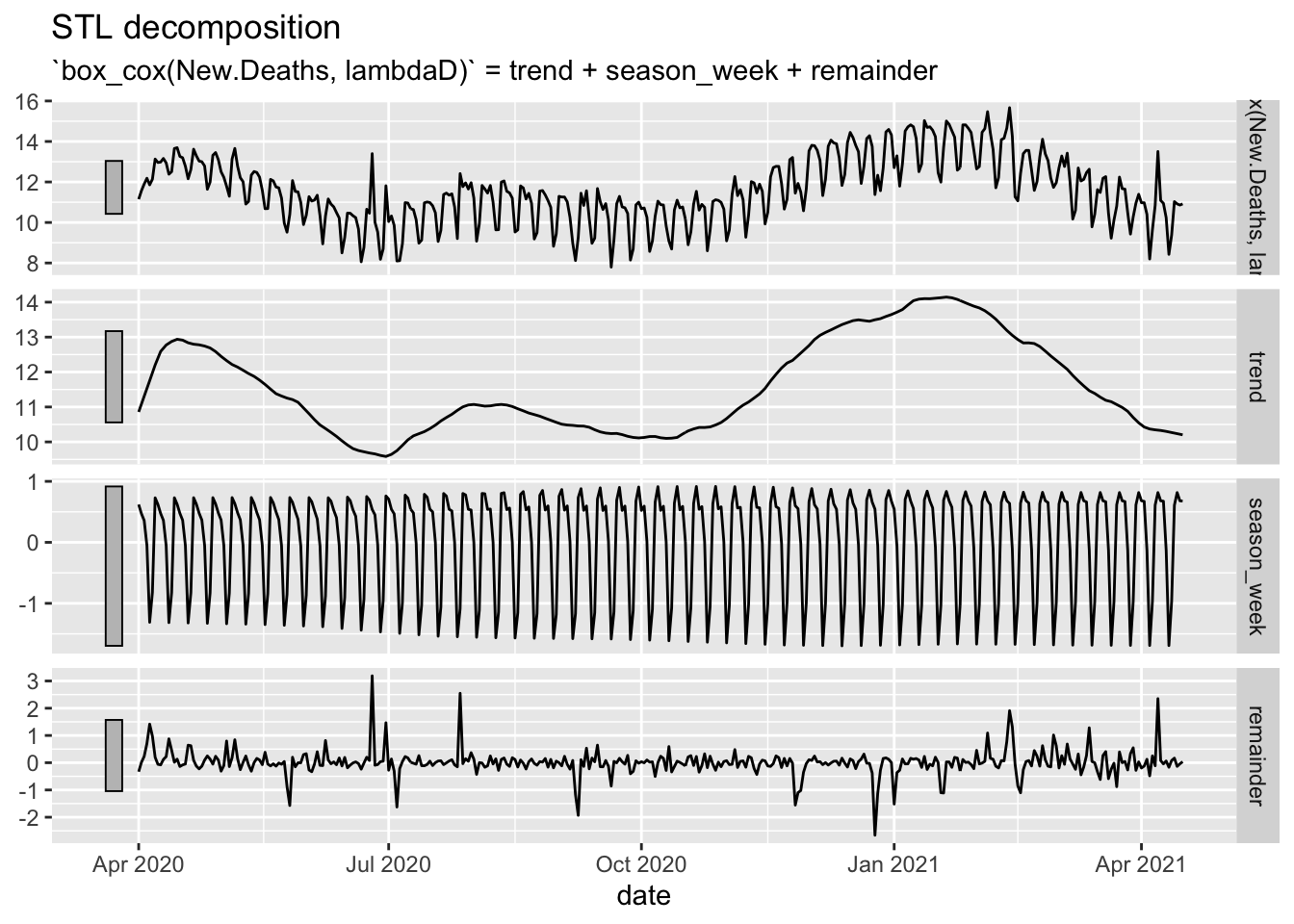
Training and Testing
That gives me the data that I want; now let me slice it up into training and test sets.
COVID.Agg.Test <- NYTAgg.COVID %>%
as_tibble() %>%
group_by(state) %>%
filter(date > as.Date("2021-03-31")) %>%
ungroup() %>%
as_tsibble(index = date, key = state)
COVID.Agg.Train <- anti_join(NYTAgg.COVID, COVID.Agg.Test) %>%
as_tsibble(index = date, key = state)Model Fitting
Fit some models to the data.
COVID.Models <- COVID.Agg.Train %>%
filter(is_aggregated(state)) %>%
model(`K = 2` = ARIMA(box_cox(New.Cases, lambdaC) ~ fourier(K = 2) + PDQ(0, 0,
0)), ARIMA = ARIMA(box_cox(New.Cases, lambdaC)), ETS = ETS(box_cox(New.Cases,
lambdaC)), NNET = NNETAR(box_cox(New.Cases, lambdaC)), NNETX = NNETAR(box_cox(New.Cases,
lambdaC) ~ Day.of.Week), ARIMAX = ARIMA(box_cox(New.Cases, lambdaC) ~ Day.of.Week),
ETSlog = ETS(log(New.Cases + 1)), ETSLevel = ETS(New.Cases)) %>%
mutate(Combo1 = (`K = 2` + ARIMA + ETS)/3, Combo2 = (`K = 2` + ARIMA + ETS +
NNET)/4, Combo3 = (NNETX + ARIMAX)/2, Combo4 = (ARIMAX + ETS)/2, Combo5 = (ETS +
ETSlog + ETSLevel)/3)Forecast the models.
COVIDFC <- COVID.Models %>%
filter(is_aggregated(state)) %>%
forecast(h = 14, new_data = COVID.Agg.Test)COVIDFC %>%
filter(is_aggregated(state)) %>%
accuracy(COVID.Agg.Test %>%
filter(is_aggregated(state)))# A tibble: 13 x 11
.model state .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr*> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 ARIMA <aggregat… Test 2468. 6554. 5297. 1.82 8.32 NaN NaN -0.0535
2 ARIMAX <aggregat… Test 271. 7164. 6030. -1.80 10.1 NaN NaN -0.0579
3 Combo1 <aggregat… Test 947. 6607. 5306. -0.657 8.83 NaN NaN -0.186
4 Combo2 <aggregat… Test 1439. 7411. 6150. -0.294 10.2 NaN NaN -0.0978
5 Combo3 <aggregat… Test NaN NaN NaN NaN NaN NaN NaN NA
6 Combo4 <aggregat… Test 373. 6094. 4951. -1.33 8.26 NaN NaN -0.169
7 Combo5 <aggregat… Test 291. 5800. 4389. -1.38 7.42 NaN NaN -0.216
8 ETS <aggregat… Test 475. 5408. 4153. -0.870 6.85 NaN NaN -0.264
9 ETSLe… <aggregat… Test 1051. 6337. 4996. -0.471 8.31 NaN NaN -0.124
10 ETSlog <aggregat… Test -653. 5923. 4708. -2.81 8.03 NaN NaN -0.234
11 K = 2 <aggregat… Test -102. 8815. 7176. -2.92 12.3 NaN NaN -0.211
12 NNET <aggregat… Test 2952. 10648. 9300. 0.850 15.2 NaN NaN 0.116
13 NNETX <aggregat… Test NaN NaN NaN NaN NaN NaN NaN NA Look at the aggregates.
COVIDFC %>%
filter(is_aggregated(state)) %>%
autoplot(NYTAgg.COVID %>%
filter(date > as.Date("2021-03-01")), level = NULL)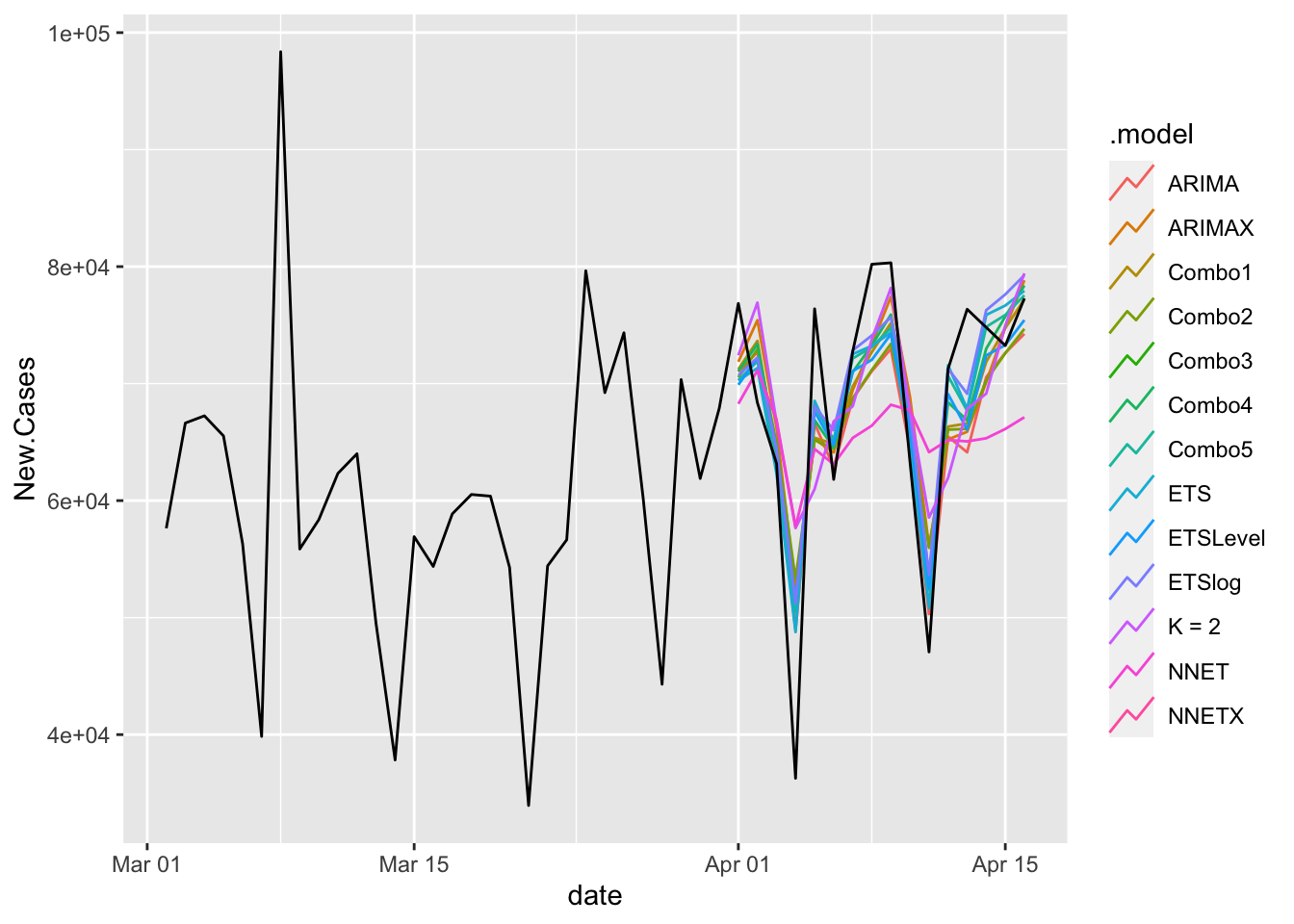
Plot the Winner
COVIDFC %>%
filter(is_aggregated(state) & .model == "ETS") %>%
autoplot(NYTAgg.COVID %>%
filter(is_aggregated(state) & date > as.Date("2021-03-01")))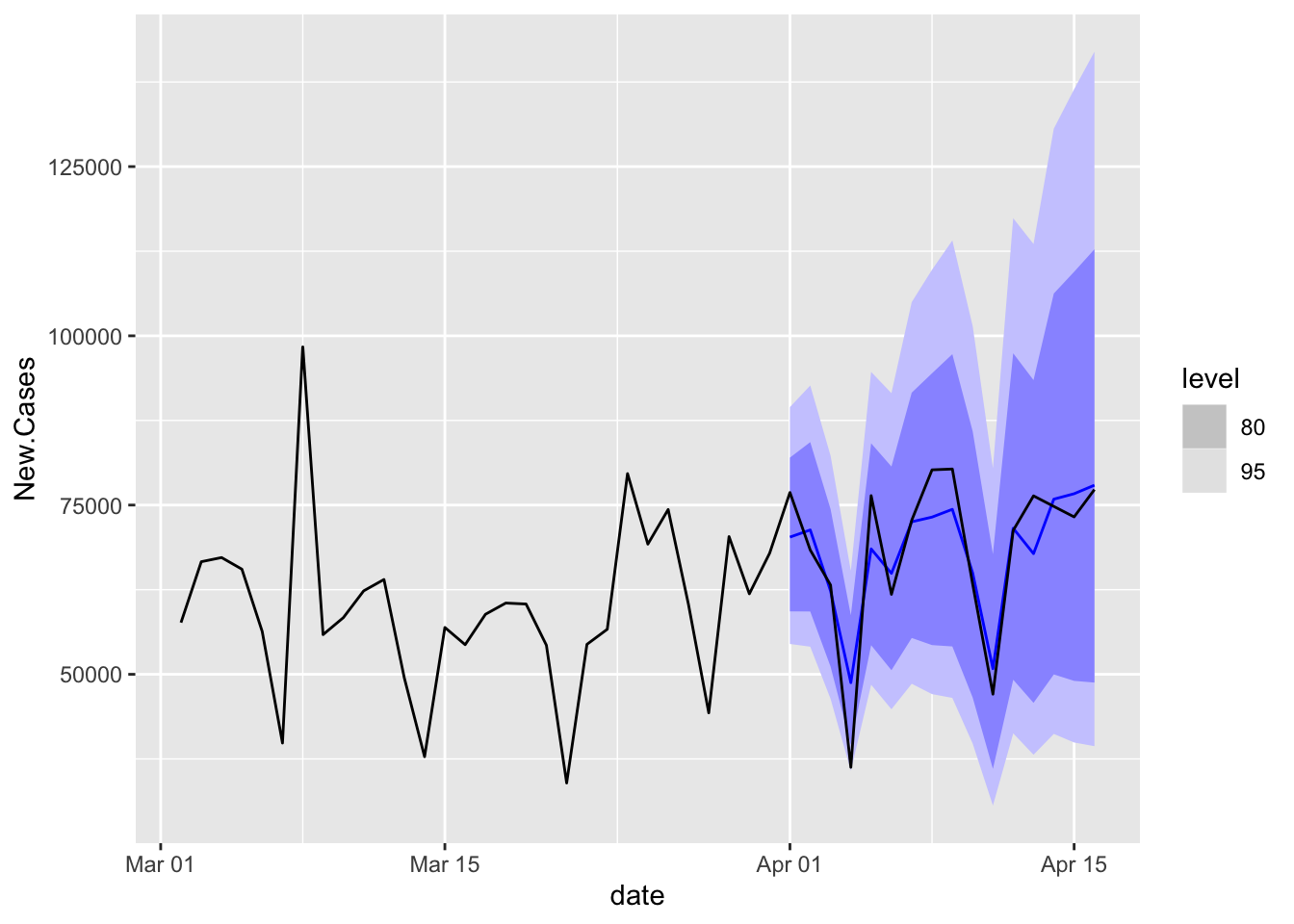
Assess the Winner
COVID.Models %>%
select(ETS) %>%
augment() %>%
ACF(.resid) %>%
autoplot()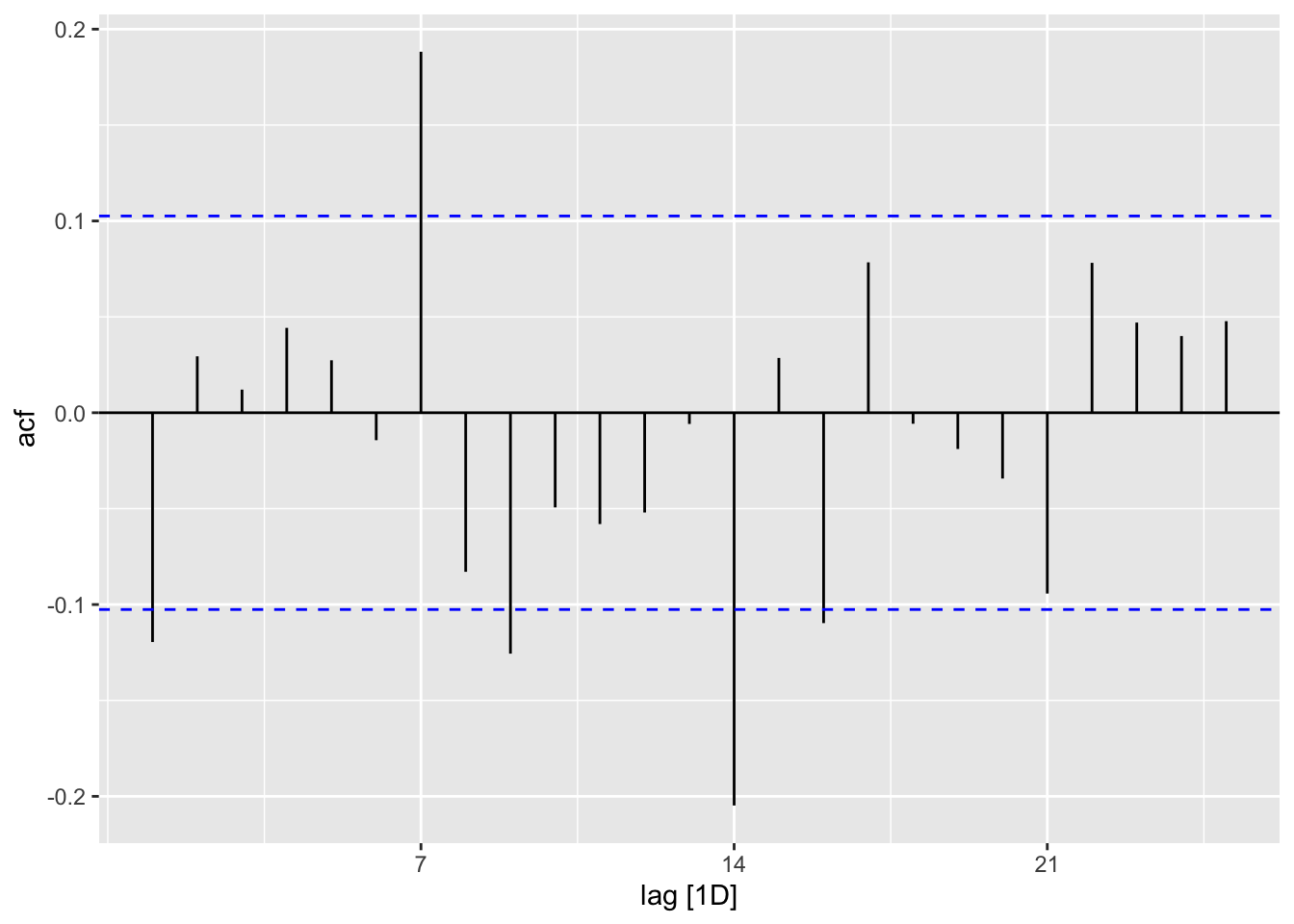
The Median Forecast?
COVID.Model.ETS <- COVID.Agg.Train %>%
filter(is_aggregated(state)) %>%
model(ETS = ETS(New.Cases))
Med.FC <- COVID.Model.ETS %>%
forecast(h = 14, point_forecast = list(.median = median), simulate = TRUE, bootstrap = TRUE,
times = 10000)
Med.FC %>%
accuracy(COVID.Agg.Test)# A tibble: 1 x 11
.model state .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr*> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 ETS <aggregated> Test 1169. 6803. 5630. -0.568 9.39 NaN NaN -0.113Med.FC %>%
autoplot(NYTAgg.COVID %>%
filter(date > as.Date("2021-03-01")))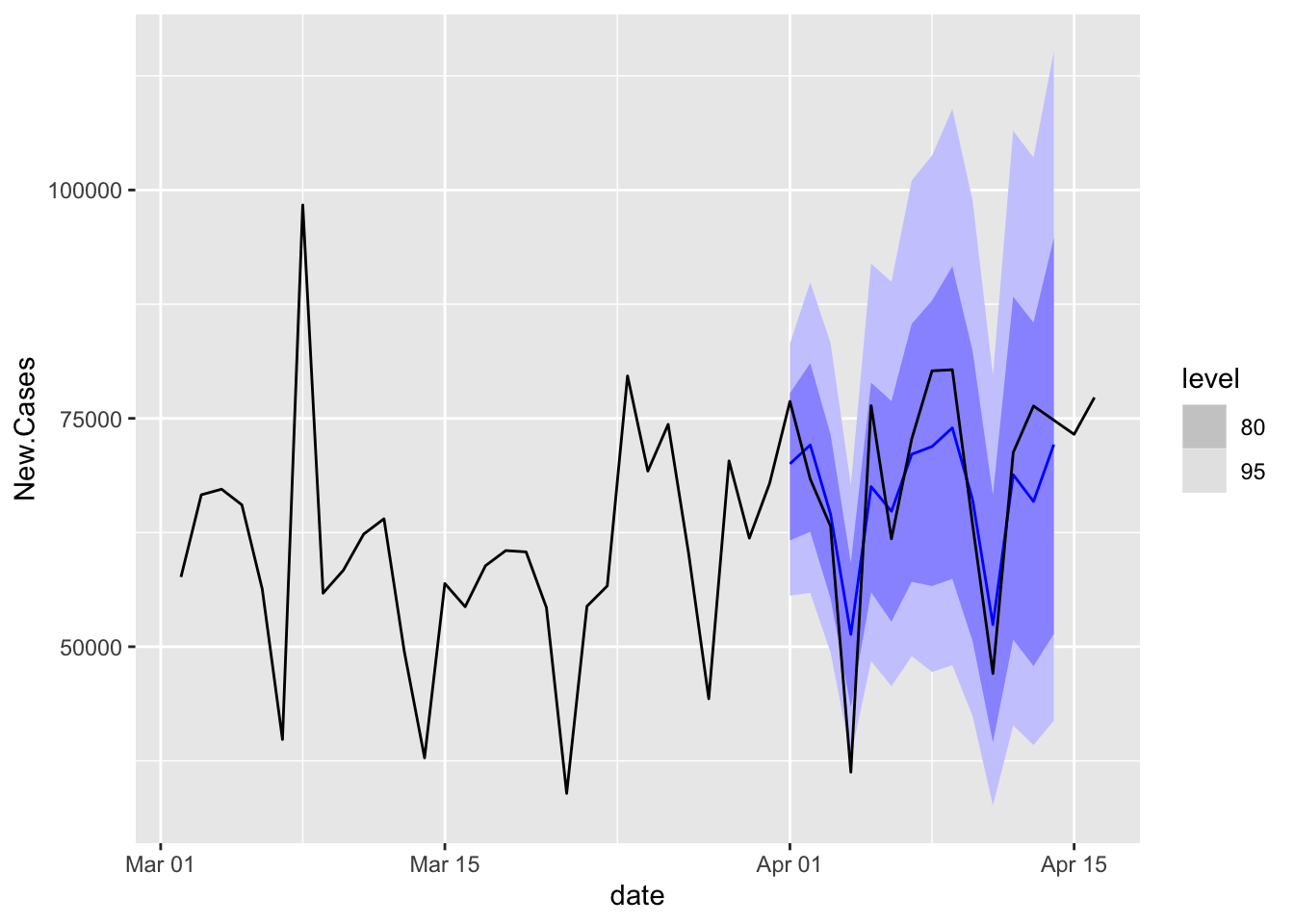
What Does the Future Hold?
ETSFA <- NYTAgg.COVID %>%
filter(is_aggregated(state)) %>%
model(ETS = ETS(New.Cases)) %>%
forecast(h = 14, point_forecast = list(.median = median), simulate = TRUE, bootstrap = TRUE,
times = 10000)
ETSFA %>%
autoplot(NYTAgg.COVID %>%
filter(date > as.Date("2021-03-01")))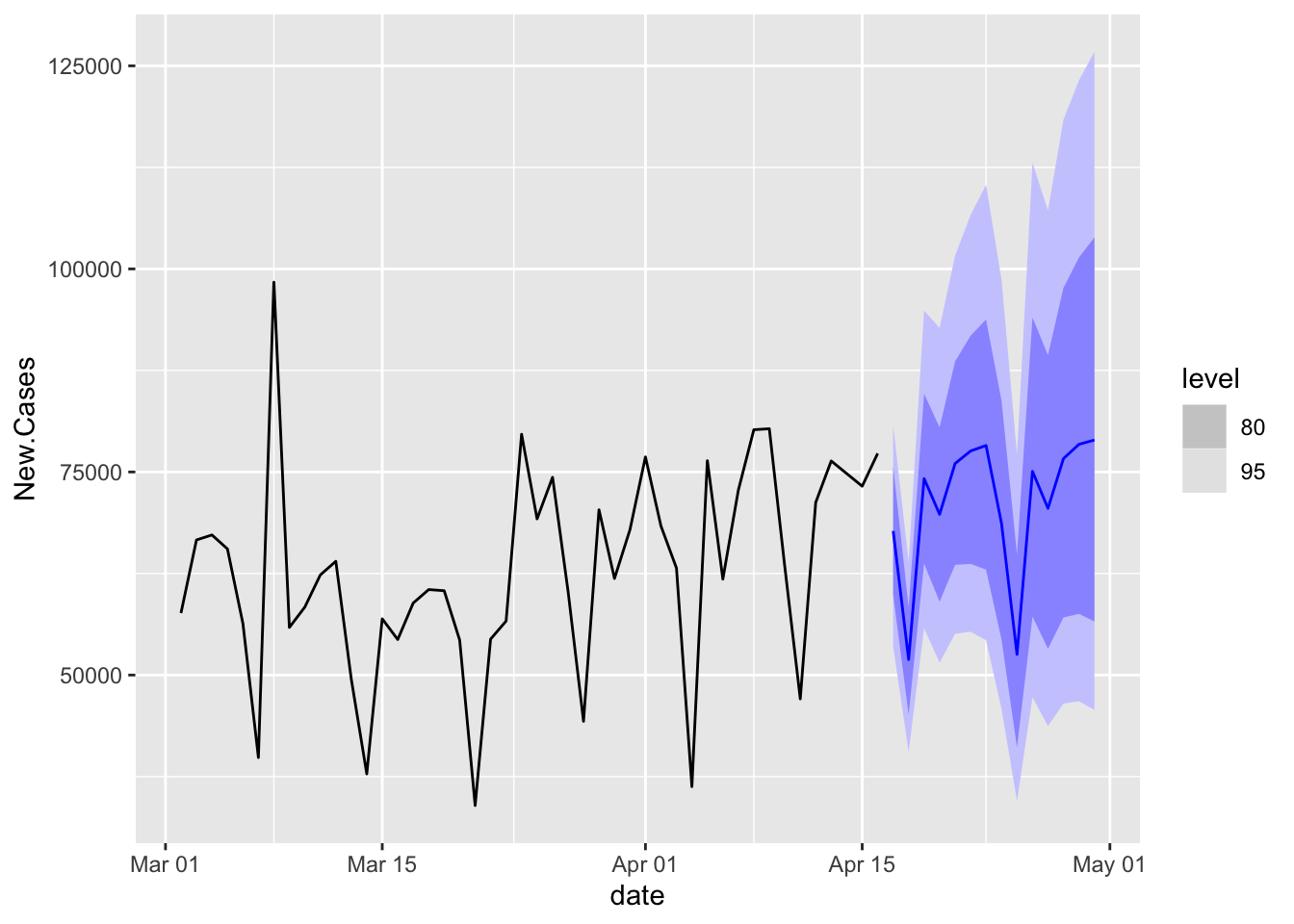
Deaths
COVID.ModelsD <- COVID.Agg.Train %>%
filter(is_aggregated(state)) %>%
model(`K = 2` = ARIMA(box_cox(New.Deaths, lambdaD) ~ fourier(K = 2) + PDQ(0,
0, 0)), ARIMA = ARIMA(box_cox(New.Deaths, lambdaD)), ETS = ETS(box_cox(New.Deaths,
lambdaD)), NNET = NNETAR(box_cox(New.Deaths, lambdaD)), NNETX = NNETAR(box_cox(New.Deaths,
lambdaD) ~ Day.of.Week), ARIMAX = ARIMA(box_cox(New.Deaths, lambdaD) ~ Day.of.Week),
ETSlog = ETS(log(New.Deaths + 1)), ETSLevel = ETS(New.Deaths)) %>%
mutate(Combo1 = (`K = 2` + ARIMA + ETS)/3, Combo2 = (`K = 2` + ARIMA + ETS +
NNET)/4, Combo3 = (NNETX + ARIMAX)/2, Combo4 = (ARIMAX + ETS)/2, Combo5 = (ETS +
ETSlog + ETSLevel)/3)Forecast the models.
COVIDFCD <- COVID.ModelsD %>%
filter(is_aggregated(state)) %>%
forecast(h = 14, new_data = COVID.Agg.Test)COVIDFCD %>%
filter(is_aggregated(state)) %>%
accuracy(COVID.Agg.Test %>%
filter(is_aggregated(state)))# A tibble: 13 x 11
.model state .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr*> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 ARIMA <aggregat… Test -95.8 390. 263. -22.7 29.3 NaN NaN -4.13e-2
2 ARIMAX <aggregat… Test -60.8 375. 231. -19.0 25.7 NaN NaN -5.67e-2
3 Combo1 <aggregat… Test -105. 389. 270. -24.0 30.4 NaN NaN -3.25e-2
4 Combo2 <aggregat… Test -63.2 396. 242. -18.3 25.3 NaN NaN -1.42e-1
5 Combo3 <aggregat… Test NaN NaN NaN NaN NaN NaN NaN NA
6 Combo4 <aggregat… Test -86.3 380. 252. -21.3 27.8 NaN NaN -5.53e-2
7 Combo5 <aggregat… Test -107. 390. 271. -23.9 30.3 NaN NaN -4.60e-2
8 ETS <aggregat… Test -112. 389. 273. -23.6 29.9 NaN NaN -4.63e-2
9 ETSLev… <aggregat… Test -76.2 378. 244. -20.6 27.1 NaN NaN -8.83e-2
10 ETSlog <aggregat… Test -132. 409. 297. -27.6 34.0 NaN NaN -3.23e-4
11 K = 2 <aggregat… Test -106. 392. 273. -25.6 32.1 NaN NaN -6.94e-3
12 NNET <aggregat… Test 61.0 480. 212. -1.19 16.2 NaN NaN -3.96e-1
13 NNETX <aggregat… Test NaN NaN NaN NaN NaN NaN NaN NA Look at the aggregates.
COVIDFCD %>%
filter(is_aggregated(state)) %>%
autoplot(NYTAgg.COVID %>%
filter(date > as.Date("2021-03-01")), level = NULL)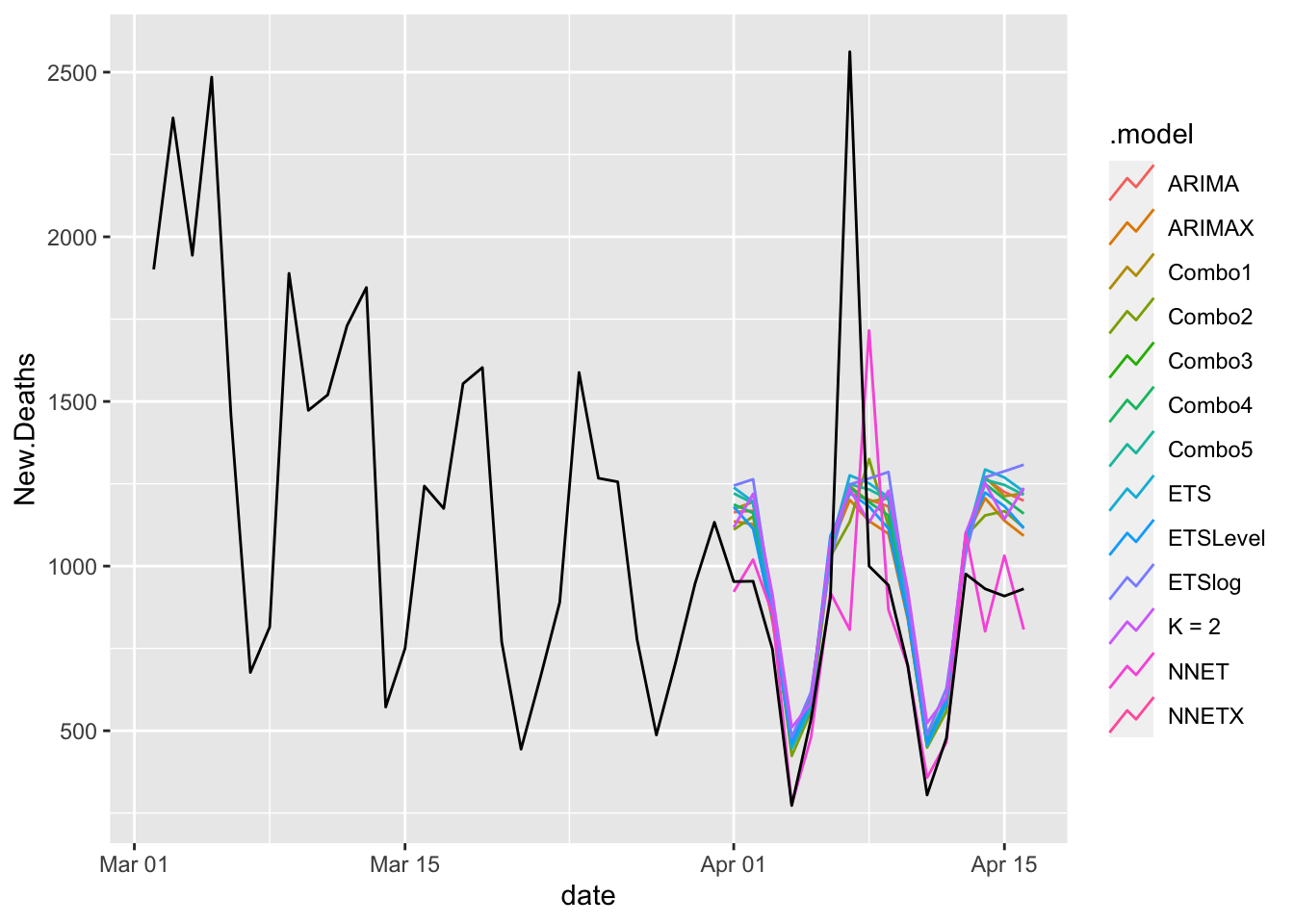 ## Diagnostics
## Diagnostics
COVID.ModelsD %>%
select(ARIMAX) %>%
report()Series: New.Deaths
Model: LM w/ ARIMA(1,1,3)(1,0,1)[7] errors
Transformation: box_cox(New.Deaths, lambdaD)
Coefficients:
ar1 ma1 ma2 ma3 sar1 sma1 Day.of.Week.L
0.0683 -0.5906 -0.1000 -0.0419 0.5341 -0.3426 1.3388
s.e. 0.4418 0.4385 0.2394 0.0936 0.3471 0.3860 0.1001
Day.of.Week.Q Day.of.Week.C Day.of.Week^4 Day.of.Week^5 Day.of.Week^6
-1.5666 -0.0362 0.3687 -0.4623 0.1221
s.e. 0.1084 0.0892 0.0831 0.0772 0.0731
sigma^2 estimated as 0.2794: log likelihood=-278.8
AIC=583.6 AICc=584.64 BIC=634.26COVID.ModelsD %>%
select(ARIMAX) %>%
augment() %>%
gg_tsdisplay(.resid)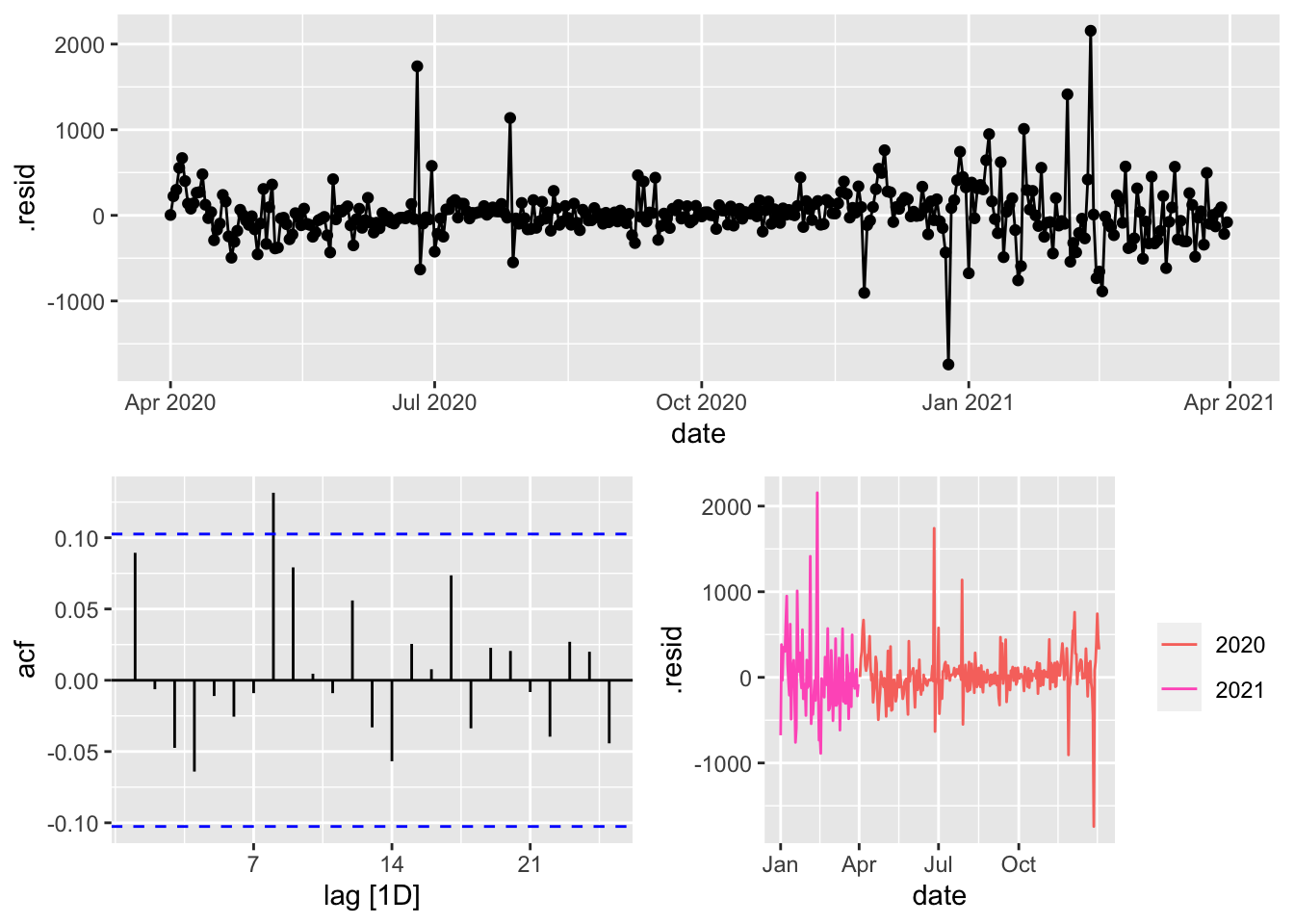
Plot the Winner
COVIDFCD %>%
filter(is_aggregated(state) & .model == "ARIMAX") %>%
autoplot(NYTAgg.COVID %>%
filter(is_aggregated(state) & date > as.Date("2021-03-01")))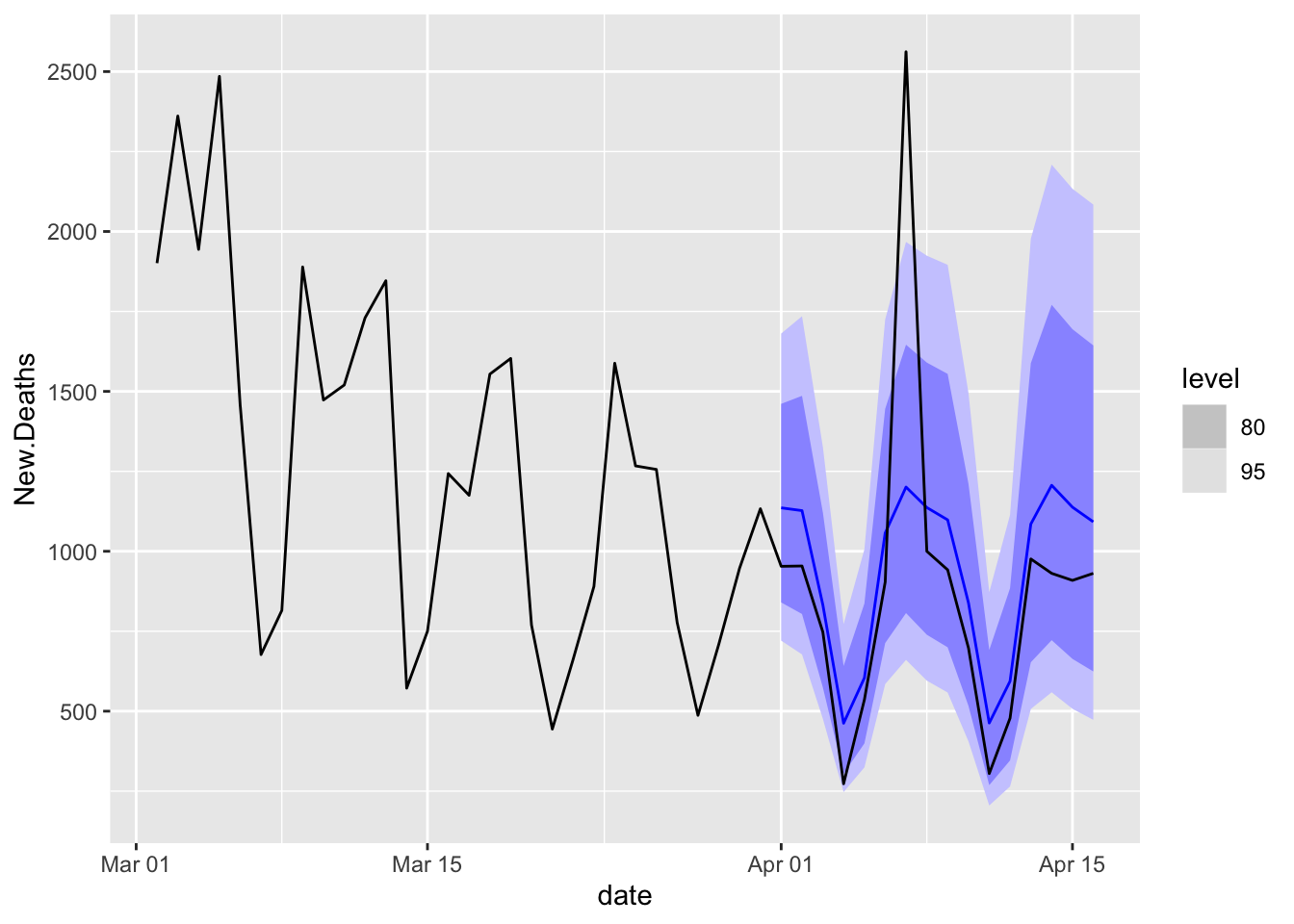
The Median Forecast?
COVID.Model.ARIMAX <- COVID.Agg.Train %>%
filter(is_aggregated(state)) %>%
model(ARIMAX = ARIMA(box_cox(New.Deaths, lambdaD) ~ Day.of.Week))
Med.D.FC <- COVID.Model.ARIMAX %>%
forecast(new_data = COVID.Agg.Test, point_forecast = list(.median = median),
simulate = TRUE, bootstrap = TRUE, times = 10000)
Med.D.FC %>%
accuracy(COVID.Agg.Test)# A tibble: 1 x 11
.model state .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr*> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 ARIMAX <aggregated> Test -58.6 374. 228. -18.8 25.4 NaN NaN -0.0603The Future
Day.of.Week <- data.frame(date = Sys.Date() + days(0:13)) %>%
mutate(Day.of.Week = wday(as.Date(date), label = TRUE)) %>%
as_tsibble(., index = date)
AggD <- NYTAgg.COVID %>%
filter(is_aggregated(state)) %>%
as_tsibble(., index = date, key = NULL)
FutureFC <- AggD %>%
model(ARIMAX = ARIMA(box_cox(New.Deaths, lambdaD) ~ Day.of.Week)) %>%
forecast(new_data = Day.of.Week, point_forecast = list(.median = median), simulate = TRUE,
bootstrap = TRUE, times = 10000)
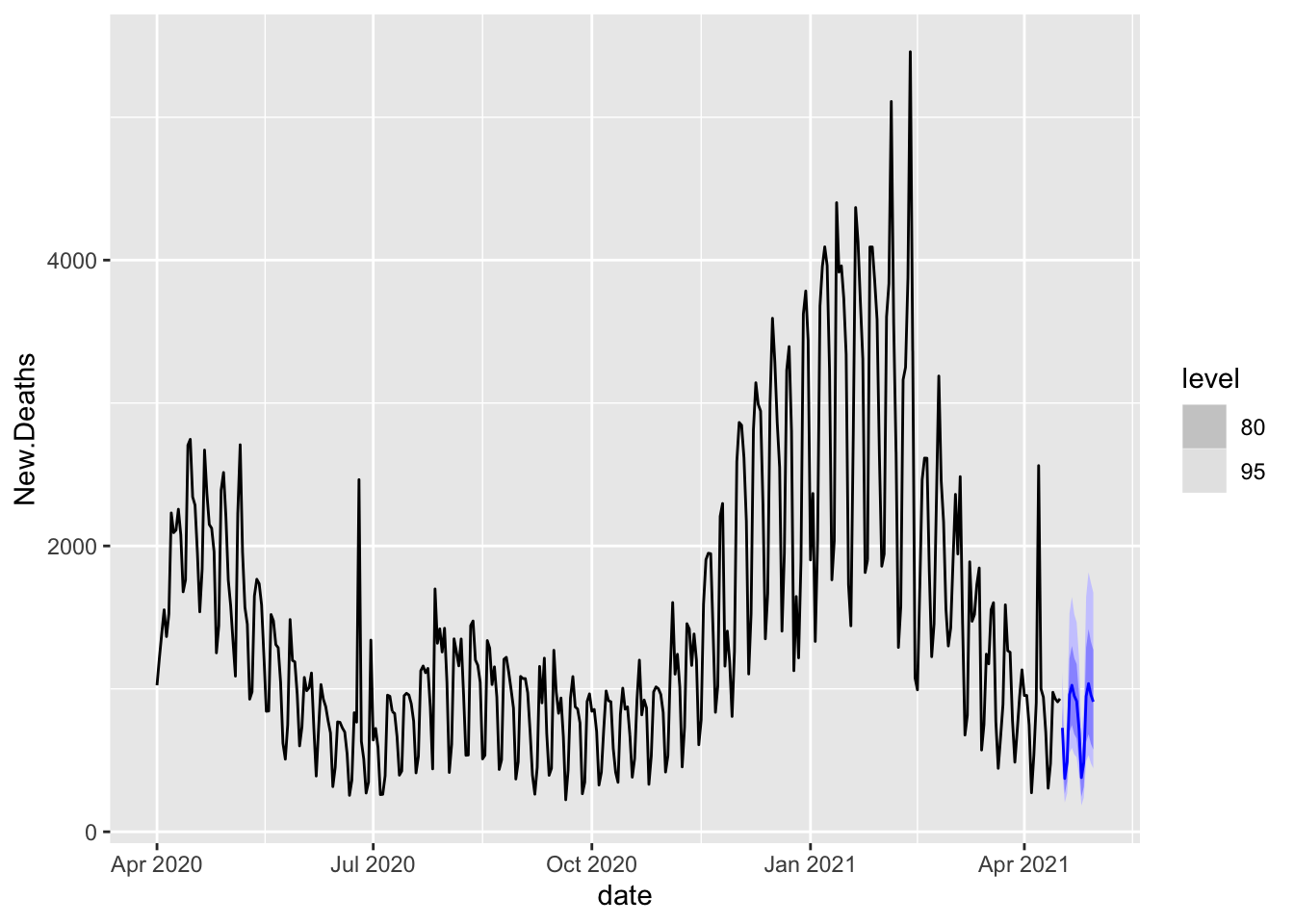
FutureFC %>%
autoplot(AggD)